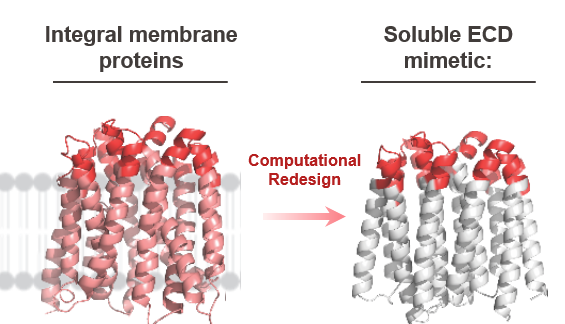
Binding proteins are proteins whose normal biological role is to bind specific molecules or ligands in cells. The research project I currently work on aims at elaborating a technique to effectively screen possible binders. It focuses on binders that interact with integral membrane proteins, that are permanently embedded in the cell membrane, such as Somatostatin Receptors . Those proteins are essential because when somatostatin binds, these receptors inhibit the release of several other hormones such as growth hormone or insulin, yet they pose an issue : their hydrophobic nature makes them difficult to handle, therefore preventing us from expressing and studying them. To overcome this problem, the LPDI has implemented a simple idea : recreate those membrane proteins into hydrophilic proteins, that preserve the initial binding capacities of the protein of interest, thus bypassing the solubility problem. To do so, they devised to redesign those membrane proteins into water-soluble mimetics computationally, by crafting a pipeline based on protein design softwares. The mimetics have the exact same extracellular domain, which is responsible for the binding capacities of the membrane protein, but an entirely new and soluble scaffold, that allows the mimetics to be highly manageable.
To confirm the efficiency of the strategy, the water soluble mimetics are now expressed experimentally, and their capacity to bind the natural ligand of the original receptor (somatostatin) will be studied. If the results show that they are indeed suitable mimetics for the original membrane proteins, they will be used to screen binder libraries to find a binder matching the receptor of interest, in this case the somatostatin receptor.
During the project, the work I will be undertaking consists mostly of the production of the desired Somatostatin Receptor mimetics, through a process called protein expression. The workflow is quite standard : order a plasmid and the desired insert (that codes for the protein), clone them into a plasmid that can then be transformed into a bacterial or mammalian cell, grow precultures and cultures of the cell that will contain the desired protein. Afterwards : lyse and destroy the cell to release the protein inside of it, discard the debris and concentrate the obtained protein solution. The expression process is now complete and is followed by the purification process, through which we make sure that we keep the protein only. It consists of a nickel affinity purification and a size exclusion chromatography. By the end of the process, we obtain a purified protein sample, whose binding properties we can determine by the use of Surface Plasmon Resonance. Firstly, we verify that the mimetics that we designed computationally will behave similarly to the original Somatostatin receptor when interacting with Somatostatin, thus confirming that the computationnal pipeline devised is suitable for the design of mimetics. Secondly, we hope to use the vetted mimetics to find suitable binders from an already existing but too consequent library of possible binders.
The results of these solubilisation efforts will therefore provide us with information about the limits of solubilizing integral membrane proteins. Development of solubilisation strategies as described in the previous sections is of major interest to pharmaceutical industry and extends our labs previously reported success in solubilisation of integral membrane proteins. The technology can make a wide range of integral membrane proteins accessible to binder design campaigns which were previously not accessible, due to their inherent characteristics that make them challenging to work with experimentally.
Please let me know about any questions or remarks you might have, I would love to discuss the matter further.


Please sign in
If you are a registered user on Laidlaw Scholars Network, please sign in
Great work, Aminata! Turning tricky, difficult to handle proteins into friendly, water loving versions sounds brilliant. It’s like giving them little swimsuits so they can play nicely in the lab. Can’t wait to see how your mimetics perform!
Thank you so much for your interest Bianca, it is a brilliant metaphor, that really sums up what we have been trying to achieve in the laboratory. I look forward to sharing the results with you !
Congratulations on your work! All of those laboratory manipulations seem so interesting — I would love to see them with my own eyes! It's incredible what we can do on such a small scale!
Thank you so much for your interest Emma ! I will try to include some pictures in my report, to better illustrate what the manipulations entail.
Fascinating work, Aminata! Excited to see where your research leads you!!
Thank you so much for your support Ben !
Thanks for this write-up! It's interesting to learn that it may be to produce mimetics of these receptors that can fulfil the same purpose, whilst also being capable of functioning in different conditions. You're doing some really amazing work!
Thank you for your attention Serena ! I have definitely discovered so much over the course of this internship, and it has helped me redefine what is or is not possible.
Super interesting Aminata ! Your approach to making complex proteins lab-friendly is impressive. Thank you for explaining the process and can't wait to see what happens next !