Preliminary Research Proposal: "Automated Procedure for the Microbial Analysis of Contact Plates"
Introduction
Research from Boyce et al. [1] among others has shown that conventional manual cleaning has significant limitations and is subject to major reliability issues. Visual inspections, which are the most common form of hygiene audits in hospitals, also have limited utility, since parthenogenic microorganisms (which may be bacteria, virus, or fungus) are typically invisible to the naked eye. As a result of these systematic issues, the prevalence of infectious disease has been steadily growing over recent decades; prior to COVID, more than 6% of patients in Irish hospitals were likely to acquire an infection during their stay [2].
Several new and exciting cleaning technologies have emerged since the start of the COVID-19 pandemic. However, to evaluate their effectiveness in real-world settings, hospitals must undertake processes of environmental testing where the bioburden on surfaces/air is analysed before and after the cleaning procedure. Such procedures place significant time and resource demands on the hospital’s microbiology department. Among the most time/resource consuming elements of this work is the counting of colony-forming units (CFUs), a task which is commonly performed 24 and 48 hours after samples are incubated. The motivation for this project is to explore how this task can be automated.
Aims
This research will focus on the automated reading of contact plates, the standard devices for environmental monitoring of surfaces and personnel in cleanrooms and medical settings. They consist of special petri plates containing suitable agar media with a raised agar surface above the rim of the plate. The goal of my research is to create a machine that can (i) take a high-resolution image of the surface of a contact plate, (ii) using artificial intelligence and computer vision techniques calculate the number of colony-forming units (CFUs) on the plate, and (iii) using aggregated data from multiple contact plates, compute the associated statistics and data visualisations.
Methodology
The creation of a working prototype that would fulfil the aims of the project can be realistically achieved in a six-week time frame. The project has four work packages. Each work package has been designed to be independent of the others. This ensures progress on each can be made concurrently, and that any delays incurred on a task only affect the work package they are in.
The first work package is to upgrade the machine for imaging the contact plates. This will involve upgrading the rig (currently a smartphone is being used to image the plates) to use a raspberry pi computer, a visual display, and a high-quality camera module.
The second work package will involve creating a labelled dataset to train a machine learning model. For this task, I will assemble a database of >1000 images of contact plates where the number of CFUs on each plate has been recorded. The data for this dataset has already been collected by the SFI 20/COV/0021 COVID Rapid Response project.
The third work package will involve evaluating the accuracy of different models for predicting the number of CFU on the plate. These include a hard-coded model and an AI-based model. My goal is to achieve an accuracy that is >95%, which is comparable with human-level performance.
The final work package will involve automatically generating statistics and graphs based on the CFU predictions. All CFU predictions will be logged in a cloud database. I will write a program that can load this data and perform significance tests, focusing specifically on the student t-test, as well as regression analysis. Associated bar plots and scatter plots will be generated too.
While it is preferable to undertake the project in Trinity, each work package can be undertaken remotely if needed due to COVID-19 restrictions.
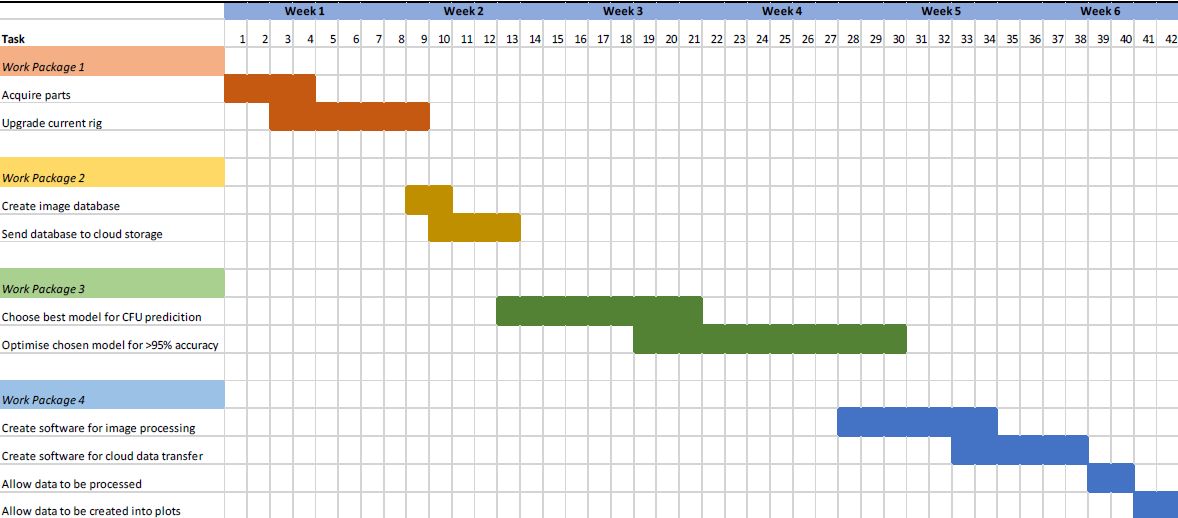 Figure 1: Preliminary Gnatt Schedule
Figure 1: Preliminary Gnatt Schedule
Interdisciplinary Opportunities
The project will contribute to ongoing research between the departments of Mechanical and Manufacturing Engineering and the Department of Microbiology. I will be cooperating closely with Microbiology researchers including Dr. Michael Beckett and Prof. Kim Roberts for work package 2.
Since the project involves the development of AI and computer vision algorithms, there is a significant computer science component to the project. Since my supervisor is a PI in the Adapt research centre, I will have the opportunity to engage with computer science experts on elements pertaining to work package 3.
Research Supervisor
I have chosen Dr. Conor McGinn as my supervisor for this research project. I have built a strong relationship with Dr. McGinn over the past two years due to him being a lecturer and a class coordinator. He is also one of the leaders of the Robotics and Innovation Lab (RAIL) at Trinity College which shows he has a great understanding of the field of robotics.
Dr. McGinn has worked closely with me to create this research proposal, as he believes that it has the potential to further aid his own research, as well as develop my presence in the robotics field. He is prepared to offer any guidance that I require and teach me any skills that are necessary to complete the project.
Outcomes and Further Collaboration
If I am successful in creating and validating the system, the project has the potential to establish a new approach for fast, reliable, low cost and safe way to measure bioburden in clinical (and other) settings. It would have the potential to be scaled quickly, enabling a practically feasible way to significantly enhance quality control levels in these settings. In the short term, my supervisor and I have deliberated on the following outputs:
- To prepare an idea disclosure document based on the design and submit to the Technology Transfer Office to evaluate if it warrants filing a patent.
- To use the device to analyse data for several upcoming research articles being developed by the PIs research group.
- To prepare an Enterprise Ireland Commercialisation Fund grant application to seek follow on funding to increase the commercial readiness of the technology beyond the project.
- To apply to the Tangent Student Accelerator based on our findings next year.
References
[1] https://aricjournal.biomedcentral.com/articles/10.1186/s13756-016-0111-x
[2] https://www.ecdc.europa.eu/sites/default/files/media/en/publications/Publications/healthcare-associated-infections-antimicrobial-use-PPS.pdf


Please sign in
If you are a registered user on Laidlaw Scholars Network, please sign in