Research Summary - Dissecting how tubulin code controls microtubule properties
Microtubules are polymers made from heterodimers of ɑ- and β-tubulins and plays a central role in the eukaryotic cell, such as mitosis, cell motility and cell signalling. It was proposed by Verhey and Gaertig (2007) that tubulin isotypes and different post translational modifications (PTM) generate a “code” that determines the structure, dynamics and mechanics of microtubules in specific biological contexts.
Studying microtubules has been challenging due to the presence of various isoforms of tubulin obtained from the mammalian brain, which undergo multiple post-translational modifications.
Using Dr. Ti’s pioneering protocol for generating recombinant human tubulin in vitro which allows the engineering of the tubulin code, it is possible to characterise tubulin with specific mutations on the tubulin code.
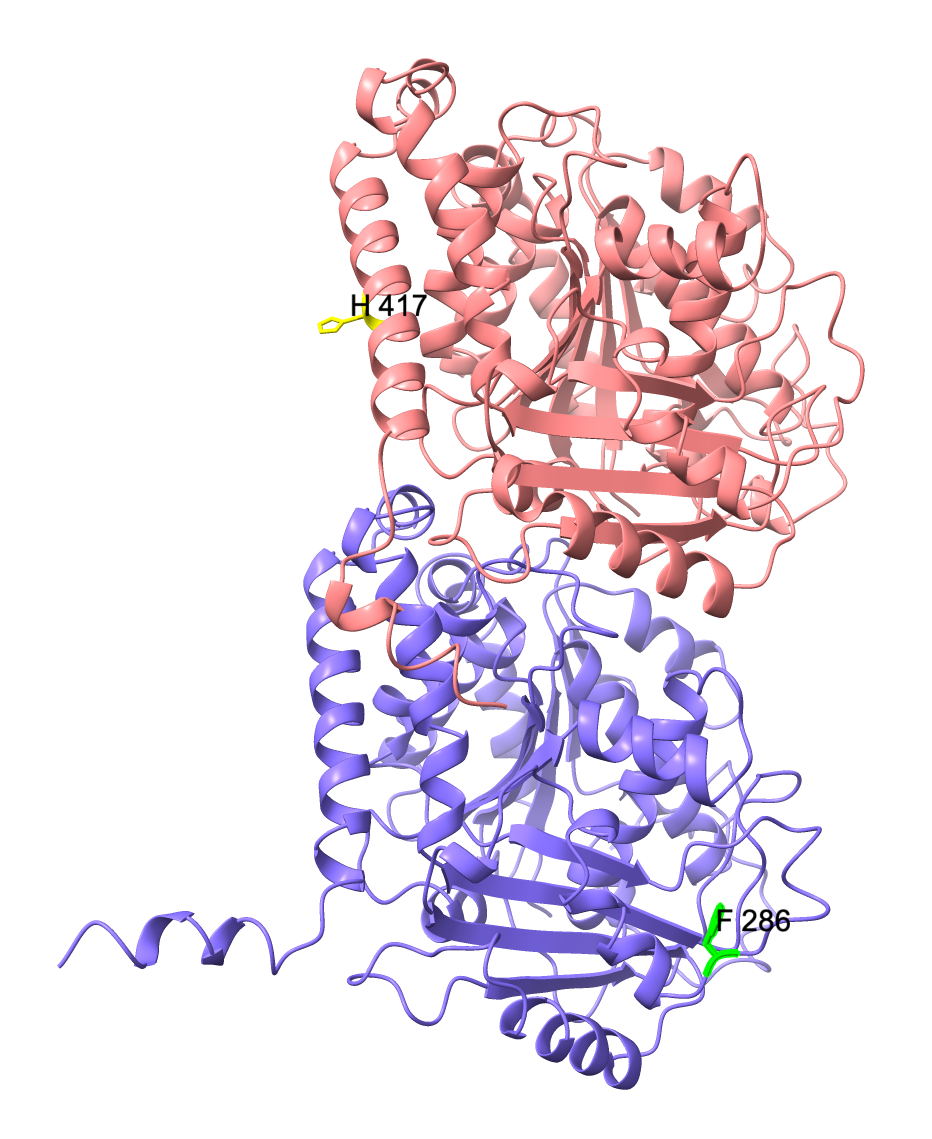
My project aims to dissect the effect of neurological disease-related mutations on microtubule properties with an initial focus on the D417H mutation as well as the L286F mutation in ɑ tubulin (TUBA1A gene), which is associated with the disease microlissencephaly (N. Bahi-Buisson et al., 2014).
Pure recombinant tubulins will be firstly generated and different isotypes will be purified using affinity chromatography, affinity tag cleavage and size exclusion chromatography, and then assembled into microtubules with Taxol (Ti. et al, 2020). Fluorescent tagged microtubules can then be imaged using TIRF microscopy and their length can be tracked to measure their polymerisation dynamics.
References
Bahi‐Buisson, N., Poirier, K., Fourniol, F. J., Saillour, Y., Valence, S., Lebrun, N., Hully, M., Bianco, C. F., Boddaert, N., Elie, C., Lascelles, K., Souville, I., Beldjord, C., & Chelly, J. (2014). The wide spectrum of tubulinopathies: what are the key features for the diagnosis? Brain, 137(6), 1676–1700. https://doi.org/10.1093/brain/awu082
Ti, S., Wieczorek, M. W., & Kapoor, T. M. (2020). Purification of Affinity Tag-free Recombinant Tubulin from Insect Cells. STAR Protocols, 1(1), 100011. https://doi.org/10.1016/j.xpro.2019.100011
Verhey, K. J., & Gaertig, J. (2007). The Tubulin Code. Cell Cycle, 6(17), 2152–2160. https://doi.org/10.4161/cc.6.17.4633



Please sign in
If you are a registered user on Laidlaw Scholars Network, please sign in