Navigating the Noise
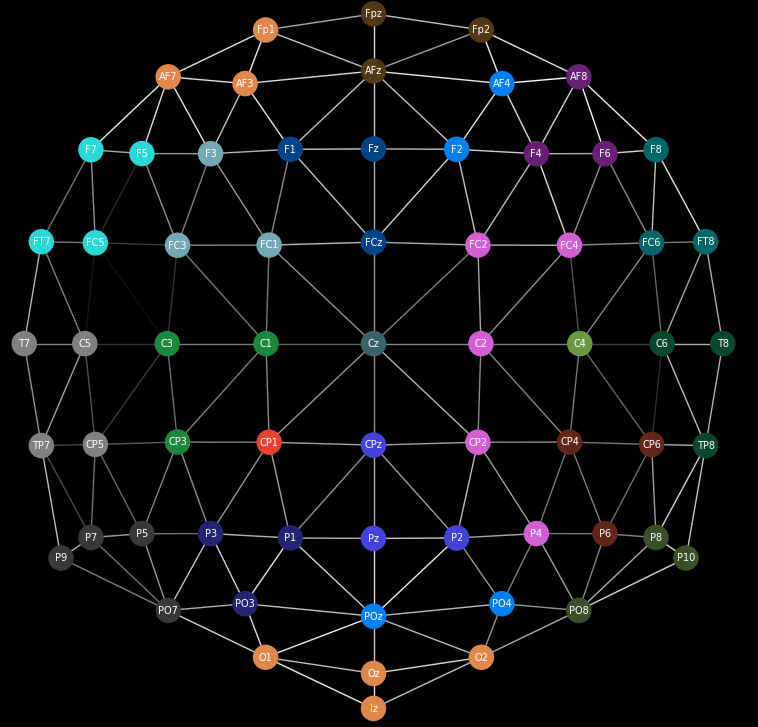
Electroencephalography (EEG) is a technique used to record electrical activity in the brain using electrodes placed on the scalp. It is a powerful, low-cost tool with high temporal resolution, meaning it can capture very fast neural activity.[1] In this project, I worked with data collected by an external collaborator at the University of Aberdeen. EEG measurements were taken while participants listened to an audiobook, under three different conditions:
- No task: Relax and listen casually (serves as a baseline)
- Semantics task: Listen carefully and prepare to answer questions based on the information provided in the audiobook afterwards
- Auditory task: Focus on the accent of the speaker and how they pronounce words (the speaker changes during the audiobook for this task)
The main goal of this research was to represent the information captured by the EEG data by modelling neural flow as a directed graph with nodes representing individual channels or brain regions. Ideally the models would show task-specific patterns across participants, and we would be able to classify which task a participant was doing by the model generated from their EEG data.
![Experimental setup during EEG acquisition [2]](https://images.zapnito.com/cdn-cgi/image/metadata=copyright,fit=scale-down,format=auto,quality=95/https://images.zapnito.com/uploads/2roTiIUFQMqAXv3fQxxU_experimental-setup-during-eeg-acquisition-and-bmi-control-a-64-channel-eeg-system-was.png)
A novel approach to this is to use Bayesian networks (BNs). A BN is a graphical representation of statistical dependence relationships among a set of variables.[3] Specifically, it is a directed acyclic graph in which nodes represent variables, and directed edges encode dependence between these variables. In this project we use dynamic BNs, in which edges only point forward in time, meaning an edge from node A to node B indicates that variable A in one moment predicts variable B a short time later. Provided with values of each variable across time, it possible to use searching algorithms to infer BNs that best capture the dependencies in the data.
When I first began this research project, I thought I had a fairly good idea of what I was getting myself into. With a background in mathematics and computer science, I was confident in my ability to tackle problems rooted in computation and statistics. What I had not anticipated was quite how messy, and unpredictable working with real EEG data would be.
The challenge wasn’t just the noise in the signal, but the sheer complexity of interpretation. Every participant, pre-processing decision, and search setting could subtly alter the resulting network, making it difficult to determine which edges genuinely carried meaning. Connections between spatially adjacent nodes couldn't be taken at face value - neighbouring electrodes often pick up overlapping signals, leading to edges that don't convey neural flow. To complicate things further, the electrodes themselves are positioned slightly differently on each subject’s scalp, introducing yet more variability.
Over time, I tried a wide range of strategies to separate meaningful information from the noise. One of my earliest experiments was the "soft overlay" of networks across participants, aiming to visualize which similar edges appeared consistently across participants for a given task. From there, I explored difference graphs to highlight changes between task and baseline conditions, experimented with separating the EEG data into specific frequency bands, and generated node degree heatmaps. I clustered nodes in hope of only retaining key information, ran statistical tests to pinpoint edges that consistently differ across conditions, and even trained machine learning classifiers to identify which graph features best distinguished between tasks.
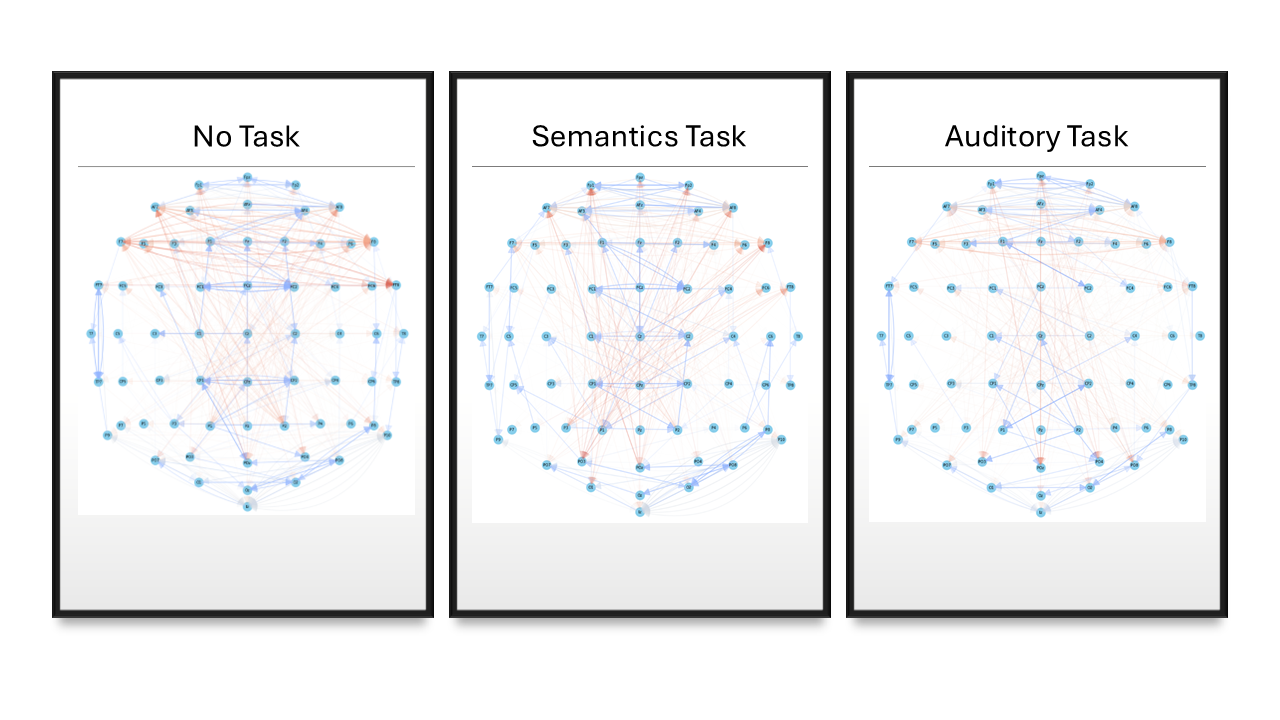
(edges between adjacent nodes are hidden)
(longer edges have a red colour to stand out)
In truth, a lot of it didn’t work - but this is where perseverance mattered most. Some of these ideas failed outright, but each helped refine my understanding and brought me a step closer to insights that felt grounded rather than speculative. I kept logs of every idea, every parameter tweak, every test performed and failed. This became invaluable. Not just for reproducibility, but for my own clarity. It was through this process - of trying, failing, reworking - that I eventually landed on a set of features and classifier that didn’t just perform well, but was interpretable, and aligned with the intuition I had started building weeks earlier.
I've thoroughly enjoyed my research experience - not only for the technical challenges it posed, but for the process of navigating uncertainty, adapting my methods, and gradually shaping a framework that brought structure to the chaos. Moving from early frustration to tangible progress has been rewarding. Looking ahead, I hope to develop this work into a publishable paper - to contribute to the field, and share approaches and insights that might support others wanting to delve deeper in the future.
References
[1] Chiarion G, Sparacino L, Antonacci Y, Faes L, Mesin L. Connectivity Analysis in EEG Data: A Tutorial Review of the State of the Art and Emerging Trends. Bioengineering (Basel). 2023 Mar 17;10(3):372. doi: 10.3390/bioengineering10030372. PMID: 36978763; PMCID: PMC10044923. [accessed 5 Jul 2025]
[2] Quantifying the role of motor imagery in brain-machine interfaces - Scientific Figure on ResearchGate. Available from: https://www.researchgate.net/figure/Experimental-setup-during-EEG-acquisition-and-BMI-control-A-64-channel-EEG-system-was_fig5_300002375 [accessed 5 Jul 2025]
[3] Hammond James and Smith V. Anne 2025 Bayesian networks for network inference in biology J. R. Soc. Interface.2220240893 https://doi.org/10.1098/rsif.2024.0893 [accessed 5 Jul 2025]


Please sign in
If you are a registered user on Laidlaw Scholars Network, please sign in